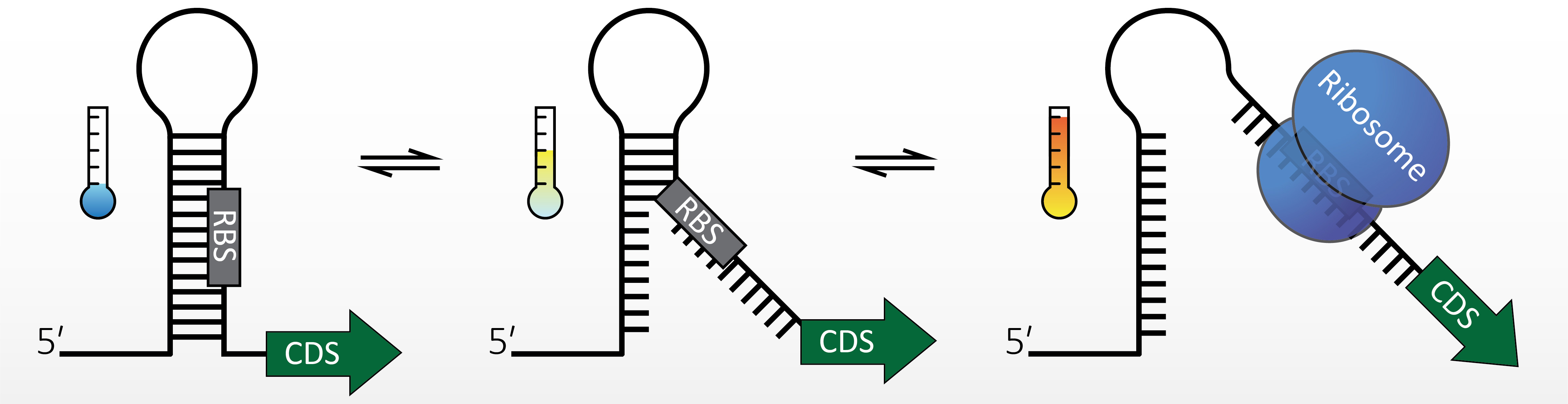
Strukturierte RNA-Segmente kontrollieren die Genexpression auf allen Ebenen. Viele bakterielle Hitzeschock- und Virulenzgene werden durch RNA-Thermometer (RNAT) reguliert. Diese molekularen Reißverschlüsse sind in der 5‘-untranslatierten Region von mRNAs lokalisiert und falten bei niedrigen Temperaturen in eine Struktur, die den Zugang des Ribosoms blockiert. Ein Aufschmelzen der Sekundärstruktur bei einem Temperaturanstieg auf 37°C (Virulenzgene) oder höher (Hitzeschockgene) legt die Ribosomen-Bindestelle frei und erlaubt dadurch die Initiation der Translation.
Die FtsH-Protease aus E. coli kontrolliert wichtige zelluläre Prozesse, wie die Hitzeschockantwort und die Lipopolysaccharid (LPS)-Biosynthese durch kontrollierten Abbau der beteiligten Faktoren. Die Hitzeschockantwort wird durch den alternativen Sigmafaktor RpoH (Sigma32) reguliert. Bei niedrigen Temperaturen wird der Sigmafaktor mit Hilfe des DnaKJ-Chaperonsystems durch FtsH abgebaut. Die LPS-Biosynthese wird durch den Abbau des Schlüsselenzyms LpxC kontrolliert. Wir interessieren uns für die Frage, wie FtsH seine Substrate erkennt und identifizieren die beteiligten Regionen in RpoH und LpxC mit Hilfe von genetischen und biochemischen Ansätzen.
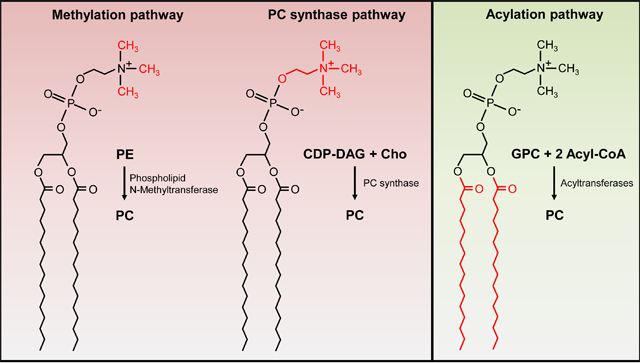
Jede lebende Zelle ist von mindestens einer Membran umgeben, die eine Barriere darstellt und vor äußeren Einflüssen schützt. Wir interessieren uns insbesondere für die Biosynthese des Membranlipids Phosphatidylcholin (PC), das nur in wenigen Bakterien vorkommt, dort aber eine wichtige Rolle in der Stressresistenz und Bakterien-Wirtsinteraktion spielt. Wir untersuchen die Enzyme von drei verschiedenen PC-Biosynthesewegen, die entweder die Kopfgruppe modifizieren oder Fettsäureketten anhängen.
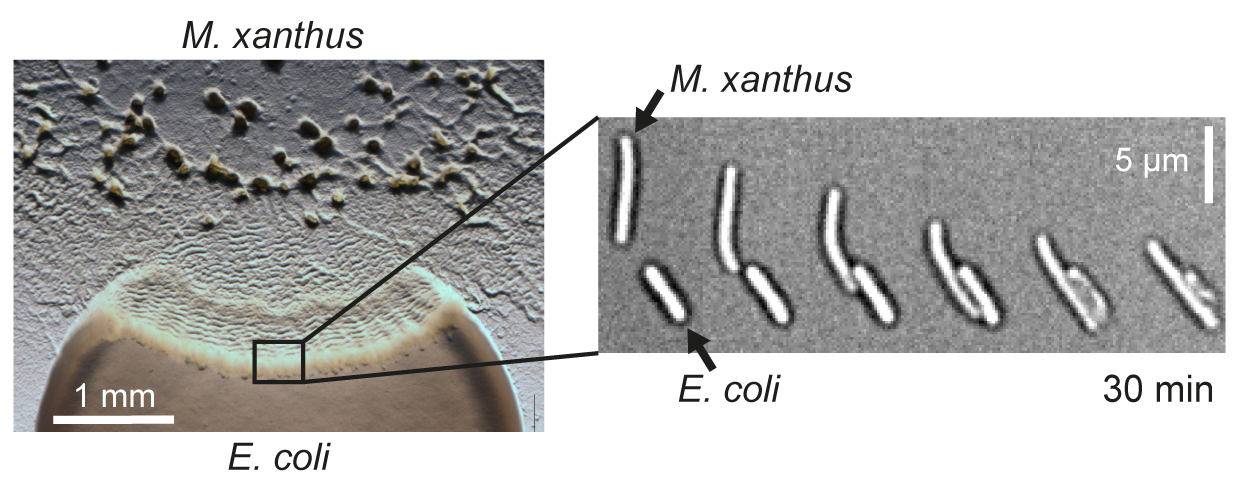
Räuber-Beute-Beziehungen spielen nicht nur bei höheren Tieren eine wichtige Rolle, sondern werden auch als Überlebensstrategie bei Bakterien beobachtet. Sogenannte „bakterielle Prädatoren“ töten gezielt Bakterien einer anderen Art ab, um deren Biomasse als Nährstoffquelle zu nutzen. Wir erforschen die molekularen Mechanismen, welche die spezifische Erkennung von Beutebakterien und deren Abtöten vermitteln, am Beispiel des sozialen Bodenbakteriums Myxococcus xanthus.
Welcome to the RNA-SURIBA homepage!
The database RNA-SURIBA contains RNA-secondary structure predictions and a set of additional
parameters for all upstream regions of all genes of diverse bacterial genomes.
It can be downloaded free of charge.
Details of the database are published in the article ‘Genome-wide bioinformatic prediction and
experimental evaluation of potential RNA thermometers’
by Waldminghaus, T., Gaubig, L. C., and Narberhaus, F.; Molecular Genetics and Genomics, 2007.
For other articles related to the database see publications.
Download options are specified in the manual.
Any comments and questions should be directed to E-Mail.
Please contact us by mail and we will provide you with the datafiles.
Narberhaus, F., Waldminghaus, T., and Chowdhury, S. (2006) RNA thermometers. FEMS Microbiol Rev 30: 3-16.
Waldminghaus, T., Heidrich, N., Brantl, S. and Narberhaus, F. (2007) FourU – A novel type of RNA thermometer in Salmonella. Mol Microbiol 65(2): 413-24.
Waldminghaus, T., Fippinger, A., Alfsmann, J., and Narberhaus, F. (2005) RNA thermometers are common in alpha - and gamma -proteobacteria. Biol Chem 386: 1279-1286.
RNA Structures od untranslated Regions in Bacteria
Database description
The database RNA-SURIBA contains RNA-secondary structure predictions and a set of additional parameters for all upstream regions of all genes from 62 bacterial genomes.
Construction and content of the database RNA-SURIBA are described in detail in the article: Genome-wide bioinformatic prediction and experimental evaluation of potential RNA thermometers by Waldminghaus, T., Gaubig, L. C., and Narberhaus, F.; Molecular Genetics and Genomics, 2007.
RNA-SURIBA contains 62 tables, one for each genome. Tables are named according to the bacterial species (see table 1). Each table consists of 36 colums with the structure predictions, sequences, gene annotation data and calculated parameters for each structure (see table 2 for details).
Download RNA-SURIBA
We provide the database RNA-SURIBA in two different formats. First, a SQL-DUMP that allows the simple use of RNA-SURIBA with any SQL-server independently of the platform. Second, ASCII flat files of each table which provide the possibility to download only the dataset of interest and not the whole database. The order of rows in the flat files is consistent with the order in table 2.
Example for installing RNA-SURIBA from the DUMP-file on a postgresql system:
In addition to the database, drawings of the structures in GIF-format of each structure prediction are provided. The drawings are stored in one zip-file for every genome named after the corresponding genome (see table 1). The files are stored in folders named after the corresponding genome (see table 1). Each file is designated according to the start of the gene on the chromosome (column 2 in each RNA-SURIBA table) in combination with the folding number (column 6 in each RNA-SURIBA table).
Example:
For gene thrL from E. coli the gene start is 190 and structure prediction with the lowest energy of -11.0 represents folding 1. The corresponding file is stored as EC/Foldings/190_1.gif.
Table 1: Genomes included in the database RNA-SURIBA
| Bacterial species | Abbreviation in RNA-SURIBA |
| Acinetobacter sp. ADP1 | AC |
| Agrobacterium tumefaciens C58 | AT |
| Aquifex aeolicus VF5 | AA |
| Bacillus cereus ATCC 14579 | BC |
| Bacillus halodurans C-125 | BAH |
| Bacillus subtilis 168 | BS |
| Bartonella henselae str. Houston-1 | BH |
| Bifidobacterium longum NCC2705 | BL |
| Bordetella pertussis Tohama I | BP |
| Borrelia burgdorferi B31 | BB |
| Bradyrhizobium japonicum USDA 110 | BJ |
| Brucella melitensis 16M | BM |
| Buchnera aphidicola str. APS | BA |
| Campylobacter jejuni subsp. jejuni NCTC 11168 | CJ |
| Candidatus Blochmannia floridanus | CBF |
| Caulobacter crescentus CB15 | CC |
| Chlamydia muridarum | CM |
| Chlamydophila pneumoniae TW-183 | CP |
| Clostridium acetobutylicum ATCC 824 | CA |
| Corynebacterium glutamicum ATCC 13032 | CG |
| Coxiella burnetii RSA 493 | CB |
| Deinococcus radiodurans R1 | DR |
| Desulfovibrio vulgaris subsp vulg. str. Hildenborough | DV |
| Enterococcus faecalis V583 | EF |
| Erwinia carotovora subsp. atroseptica SCRI1043 | ER |
| Escherichia coli K12 | EC |
| Gloeobacter violoceus PCC7421 | GV |
| Haemophilus influenzae Rd KW 20 | HI |
| Helicobacter pylori 26695 | HP |
| Lactobacillus plantarum WCFS1 | LP |
| Lactococcus lactis subsp. lactis Ill403 | LL |
| Listeria monocytogenes | LM |
| Mesorhizobium loti | ML |
| Mycobacterium tuberculosis H37 Rv | MT |
| Mycoplasma genitalium G-37 | MG |
| Neisseria meningitidis MC58 | NM |
| Nitrosomonas europaea ATCC19718 | NE |
| Pasteurella multocida subsp. multocida str. Pm70 | PM |
| Photorabdus luminescens subsp. lumondii TT01 | PL |
| Pseudomonas aeroginosa PAO1 | PA |
| Pyrococcus furiosus DSM 3638 | PF |
| Ralstonia solanacearum GMI1000 | RS |
| Rhodopseudomonas palustris CGA009 | RHP |
| Ricketsia prowazekii str. MadridE | RP |
| Rickettsia conorii str Malish 7 | RC |
| Salmonella enterica subsp. enterica serovar Thphi str.CT18 | SE |
| Shewanella oneidensis MR-1 | SO |
| Shigella flexneri 2a str. 301 | SF |
| Sinorhizobium meliloti | SM |
| Staphylococcus aureus subsp. aureus MW2 | SA |
| Staphylococcus epidermidis ATCC 12228 | STE |
| Streptococcus pneumoniae R6 | SP |
| Streptomyces coelicolor A3(2) | SC |
| Synechocystis sp. PCC6803 | SS |
| Thermus thermophilus HB27 | TT |
| Treponema deticola ATCC 35405 | TD |
| Vibrio cholerae 01 biovar eltor str. N16961 | VC |
| Wigglesworthia glossinidia | WG |
| Wolbachia endosymbiont of Drosophila melanogaster | WE |
| Xanthomonas axonopodis pv. citri str. N16961 | XA |
| Xylella fastidiosa 9a5c | XF |
| Yersinia pestis CO92 | YP |
Table 2: Parameters contained in the database RNA-SURIBA
| Column in database | Name of variable | Data type | Description |
| 1 | geneName | string | Gene name according to genome annotation. |
| 2 | geneStart | int | Gene start according to genome annotation. |
| 3 | geneEnd | int | Gene end according to genome annotation. |
| 4 | geneProduct | string | Gene product according to genome annotation. |
| 5 | ene rgy | double | Free energy of RNA secondary structure prediction with mfold. |
| 6 | folding | int | Numbers indicate suboptimal RNA structure predictions (1: prediction with lowest Δ G, 2 and 3: second and third best predictions). |
| 7 | ggagg | boolean | Is the sequence “ggagg” (ideal Shine-Dalgarno) contained in the last 30 nucleotides upstream of the 3’-end? |
| 8 | plus | boolean | True, if sequence corresponds to the plus-strand. |
| 9 | aContent | float | Content of adenin in the sequence (%). |
| 10 | cContent | float | Content of cytosine in the sequence (%). |
| 11 | gContent | float | Content of guanine in the sequence (%). |
| 12 | uContent | float | Content of uracil in the sequence (%). |
| 13 | endLoopNumber | int | Number of end-loops in the structure. |
| 14 | endLoop4Number | int | Number of tetra-loops in the structure. |
| 15 | endLoop5Number | int | Number of penta-loops in the structure. |
| 16 | bulgeNumber | int | Number of bulges in the structure. |
| 17 | internalLoopNumber | int | Number of internal loops in the structure. |
| 18 | joinNumber | int | Number of joins in the structure. |
| 19 | componentNumber | int | Number of substructures in the structure. |
| 20 | basePairNumber | int | Number of base-pairs in the structure. |
| 21 | endLoopSize | float | Average size of end-loops. |
| 22 | internalLoopSize | float | Average size of internal loops. |
| 23 | bulgeSize | float | Average size of bulges. |
| 24 | endLoopBases | float | Number of nucleotides in end-loops (%). |
| 25 | internalLoopBases | float | Number of nucleotides in internal loops (%). |
| 26 | bulgeBases | float | Number of nucleotides in bulges (%). |
| 27 | stackBases | float | Number of nucleotides in stacks (%). |
| 28 | joinBases | float | Number of nucleotides in joins (%). |
| 29 | externalBases | float | Number of external nucleotides at the 3'- and 5'-end plus intern external nucleotides (%). |
| 30 | internExternalNumber | int | Number of unpaired nucleotides on the baseline (not in loops or joins and not at the 3'- and 5'-end). |
| 31 | external5Number | int | Number of external nucleotides at the 5'-end. |
| 32 | external3Number | int | Number of external nucleotides at the 3'-end. |
| 33 | loopDegree | float | Average loop-degree (How many stacks originate from one loop?). |
| 34 | seq | string | 110 nucleotides of genes (AUG +4 nucleotides of the coding region and 103 nucleotides upstream of AUG). |
| 35 | struct | string | Structure prediction in dot-bracked annotation. |
| 36 | connectStruct | string | Structure prediction in graph-annotation, corresponding to ct-file from mfold output. |